What can you learn from this article?
- Understand the concepts of data linkage, especially deterministic linakge.
- Address linkage error in the conjunction of MIMIC III (served in a postgreSQL database) and ODK database.
- Employ R to design the Extract, Transform, and Load (ETL) pipeline.
- Use Quarto document to generate a report in PDF format.
Concepts of Data Linkage
In a data scientist’s typical day, the merge/join function is an inevitable task. Data quality must be secured through linkage within one dataset (‘internal linkage’) or across multiple datasets.
According to the OECD Glossary of Statistical Terms, data linakge means a merging that brings together information from two or more sources of data with the object of consolidating facts concerning an individual or an event that are not available in any separate record.
In situations of uncertainty, there might be an linkage error, i.e. misidentifying relationships that belong to the same entity. Notably, linkage error doesn’t adhere to an binomial distribution (0 or 1); rather, it resembles a spectrum ranging from high agreement to high disagreement, along with matching possibilities influenced by quality of matching variables.
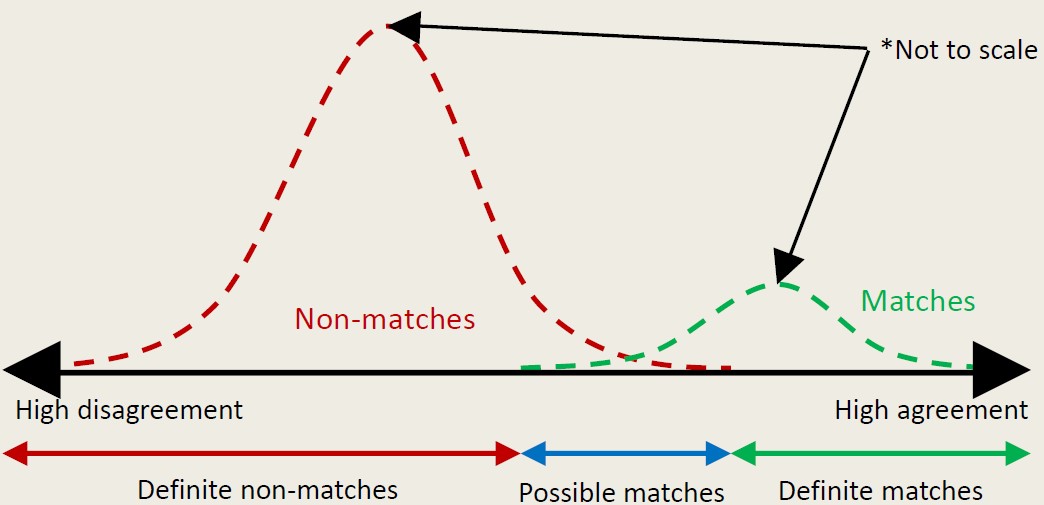 Source: James Doidge, 2023
Source: James Doidge, 2023
In order to evaluate the performance of data linkage, we can build a matrix for the link-match classification. In theory, we wish our data to achieve a ~100% sensitivity (proportion of matches that are linked), or recall, while in parralel keeping the specificity (proportion of non-matches that are not linked) high as well. To achieve these goals, three methods for data linkage are widely used, including deterministic linakge, probalistic linkage, and machine learning.
| Match (records from the same entity) | Non-match (records from different entities) | |
|---|---|---|
| Link | True match | False match |
| Non-link | Missed match | True non-match |
Source: Collaboration in Research and Methodology for Official Statistics, European Commission
I will apply the deterministic linkage method in this article. A set of predetermined rules is used to identify patterns as links or non-links. For instance, the high degree of certainty/agreement required for deterministic linkage is achieved through a unique identifier for an entity, such as NHS number. This method may allow a small amount of preconceived typographical error; however, the limitations of this method lie in the event of lower quality matching variables or handling large numbers of matching variables.
A scenario during COVID pandemic
Let’s dive into a practical scenario where data linkage proves invaluable. Suppose you are a data analyst in a UK hospital, where data is captured in the MIMIC III database and ODK database. Your hospital management has asked you to generate an executive report synthesising recent patient data from MIMIC III, survey data from ODK, and national statistics for COVID-19. Besdies, to address surges in COVID-19 cases, you should be able to regularly generate this report, with potential changes to the prototype.
What’s your strategy on this mission?
DBI
First, a database interface (DBI) is required to initialize the extract, transform, load (ETL) workflow. DBI package can help connect my R to database management systems (DBMS).
As MIMIC III is served in a PostgreSQL database, I chose RPostgres package to help me build the connection from PostgreSQL to R. Notably, RPostgres package can better capture the variables with timestmaps, comepared to the PostgreSQL package. A few lines of postgreSQL code are executed here to retrieve the specific data I need.
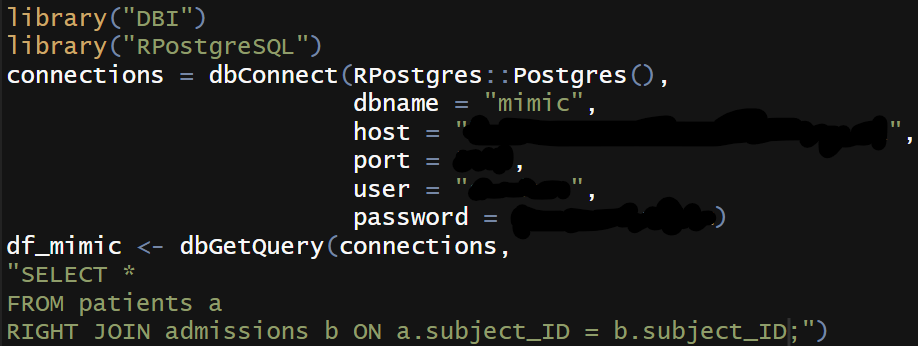
I then use ruODK package to configure and download data from ODK Central to R.
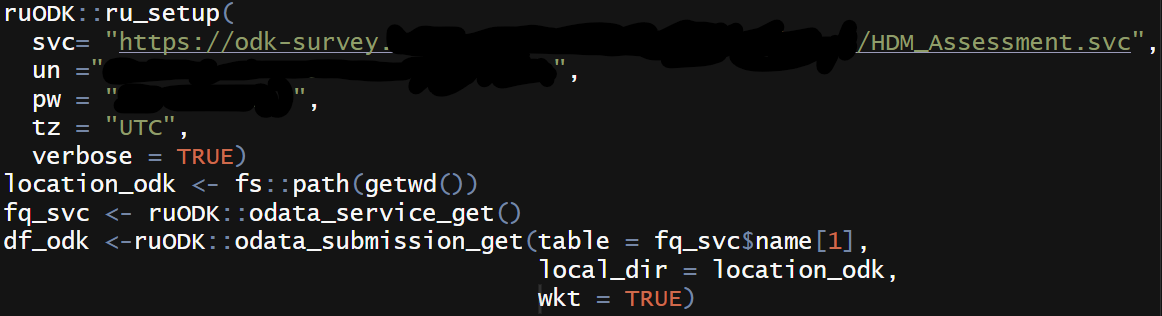
As for national statistics, I use the download.file() function in base R to get a CSV file from the data.gov.uk.
Data Linkage
Now, I are about to merge the data from MIMIC III and ODK for further cleaning, transforming, filtering, and analysis. Based on deterministic linkage method, I assume/regard the subject_id as our reliable agreement for matching between two datasets.
After utilizing R’s merge() for an inner join, I want to examine the quaity of the data linkage by introducing two additional matching variables: age at admission and gender.
Linkage error in age
Substantial variation in differnce in ages is noted following the linkage. The distribution of age difference for 25% to 75% of patients shows a deviation of within one year, which is deemed acceptable owing to rounding. Nevertheless, noticeable errors are evident in the minimum (-39) and maximum (303) age differences.
Difference in ages between MIMIC III and ODK:
| Min. | Q1 | Median | Median | Q3 | Max. |
|---|---|---|---|---|---|
| -39 | -1 | 1 | 33.95 | 1 | 303 |
To address this issue, we can exclude observations that surpass the expected lifespan. Subsequently, consult the data provider to determine which data source is more reliable regarding age entry. You can either rely on a single dataset as the definitive age or eliminate observations with significant deviations in ages.
Linkage error in gender
Let’s move on to the gender variable. There seems a bit mismatches, such as “Female-Man” and “Male-Woman”. Again, you can decide either omit all observations with mismatches in genders, or rely on a single dataset as the definitive gender.
Comparison of gender categories between MIMIC III and ODK:
| MIMIC III / ODK | Man | Woman | Trans | Non-binary |
|---|---|---|---|---|
| Female | 6 | 2620 | 4 | 4 |
| Male | 2953 | 2 | 8 | 10 |
It is also noteworthy that individuals identifying as trans and non-binary are retained in my merged data frame. This inclusion is crucial as gender minority groups may feel uncomfortable when presented with a table offering only binary options for female and male. In other words, for me, they are not linkage error; instead, ‘it’s a defect in the gender variable of MIMIC III.
After handling the linkage error, I then apply some exploratory data analysis (EDA) to privde summary satatistics for our patients. This data processing is not covered in this article.
Quarto
Finally, following data anaylsis, I use Quarto, the next generation of R Markdown released in 2022, to play as our publishing system.
You can smoothly do all the programming in a single qmd file for Quarto document. Alternatively, you can also save your output from your previous R scripts somewhere, and then open a new qmd file to read and print your outputs. Knitr::kable and kableExtra package is highly recommended for generating decent LaTeX tables in a PDF format.
Conclusion
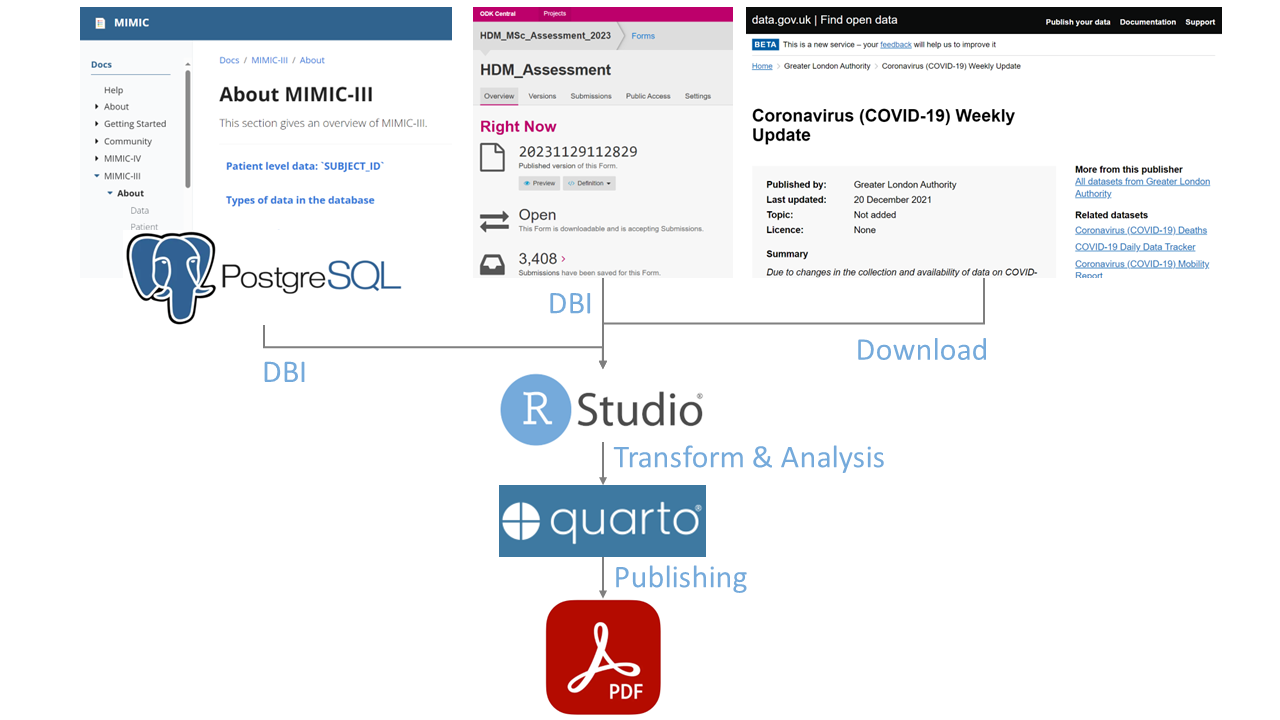
In short, investigating the matching variables may provide an indication of confidence in the link. In my scenario, I minimized data linkage error by initially agreeing on subject_id, and then stepwisely and partially agreeing on age and gender.
Additionally, processing data from various sources is achieved by using R to execute the ETL pipeline, ensuring a reproducible workflow for synthesizing a report as below.
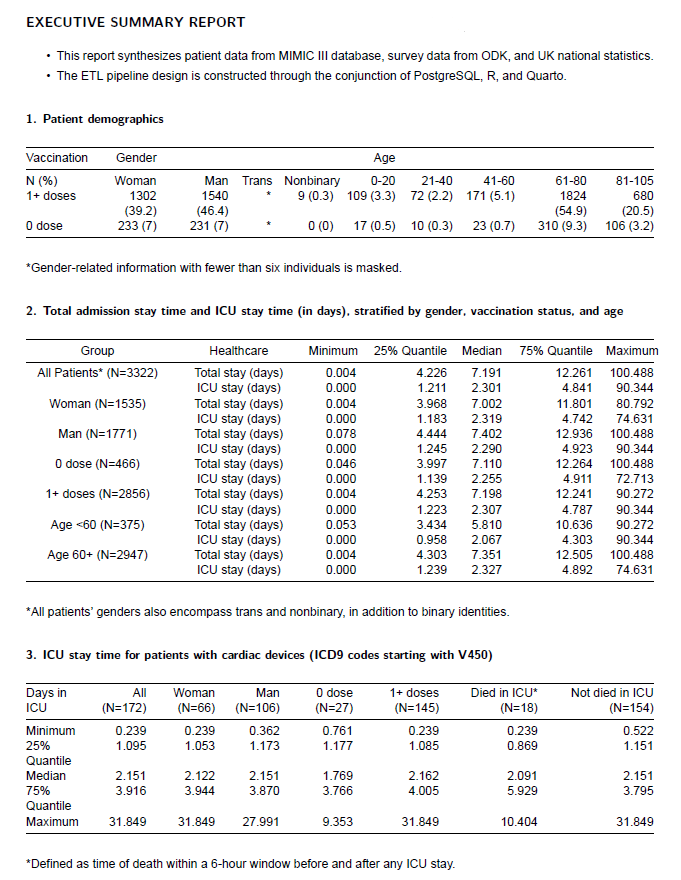
This article has been adapted from the assessment of the Health Data Management module at LSHTM. With thanks from James Doidge who introduced the data linkage topic in the Thinking Lika A Health Data Scientist module.
